normalization problem
- Read more about normalization problem
- 1 comment
- Log in or register to post comments
老师们好:
我按照严老师的dparsfa 的V4模板参数跑了rs fMRI, 没有报错。但是跑完后进行QC的时候发现有几个被试的配准特别差,如下图。我查了他们的静息态和T1都没有问题,想请问一下为什么出现这个问题?是否能纠正以及如何纠正?
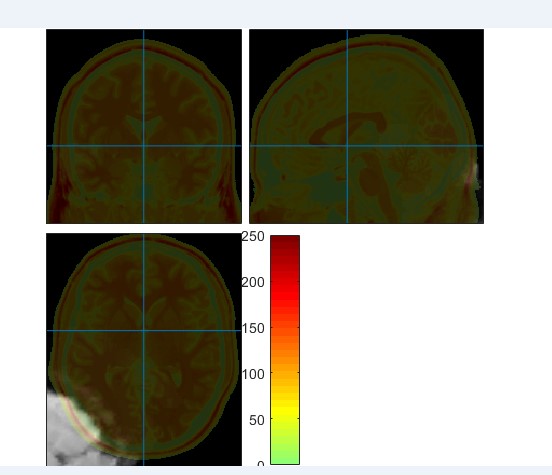
恳请老师们能予以帮助,谢谢各位老师。
老师们好:
我按照严老师的dparsfa 的V4模板参数跑了rs fMRI, 没有报错。但是跑完后进行QC的时候发现有几个被试的配准特别差,如下图。我查了他们的静息态和T1都没有问题,想请问一下为什么出现这个问题?是否能纠正以及如何纠正?

恳请老师们能予以帮助,谢谢各位老师。
Prof. Yan,
Hei, I want to consult that whether the VBM analysis of DPARSF Advanced Edition in DPABI_V4.2_190919 is based on the processing step of CAT12 of SPM12 ?
跑了一些没有T1的数据,得到了reho,falff。奇怪的是总是皮层大面积“激活”,其余部分都是“负激活”。也看不到默认网络。请问这种情况应该从哪里找原因呢。谢谢
有一些被试voxel size是1.8 1.8 5,这样的被试平滑核设置为多少才合适呢?
Dear All,
I got the error when I was performing the pre-processing for the resting data in the DPABI.
Here is the error information:
Error using DPARSFA_run (line 1620)
Index exceeds array bounds.
Error in DPARSFA>pushbuttonRun_Callback (line 1795)
[Error]=DPARSFA_run(handles.Cfg);
Error in gui_mainfcn (line 95)
Dear All,
I got the error when I was performing the pre-processing for the resting data in the DPABI.
Here is the error information:
Error using DPARSFA_run (line 1620)
Index exceeds array bounds.
Error in DPARSFA>pushbuttonRun_Callback (line 1795)
[Error]=DPARSFA_run(handles.Cfg);
Error in gui_mainfcn (line 95)
严老师好:
谢谢您的回复!
附件3是我两个被试的ARCW及Normalized 后需要检查的图像。请您看看是否有问题。谢谢!
祝好,
璐
Dear, Chaogan Group
I have some Rat data.
But I find my rat data looks upside down. I want to know when to rotate the image?
best,
Zheng Hui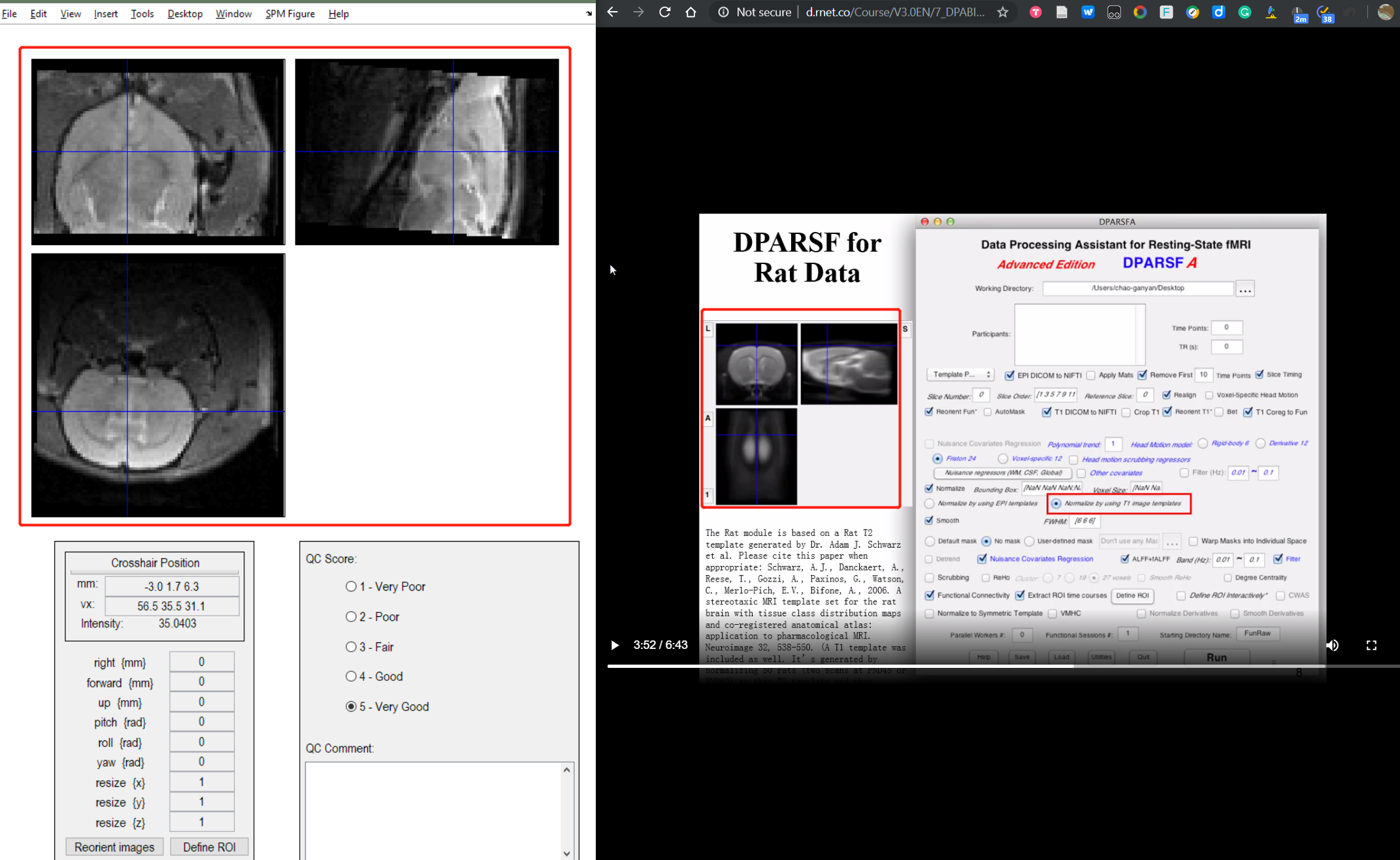
严老师,好!
Error using y_Organize_fmriprep>(parfor body) (line 203)
No matching files were found.
Error in y_Organize_fmriprep (line 201)
parfor i=1:Cfg.SubjectNum
Error in DPABISurf_run (line 479)
Hello,
我用自己的数据生成的AutoMasks显示如下,可以明显看出是有问题的。但是我用ProcessingDemoData按照同样的方式去做,显示是正常的,如下所示。我按照要求放入FunRaw和T1Raw,请问这是什么原因造成的呢?是我的数据有问题吗?有人能够帮助我吗?谢谢!