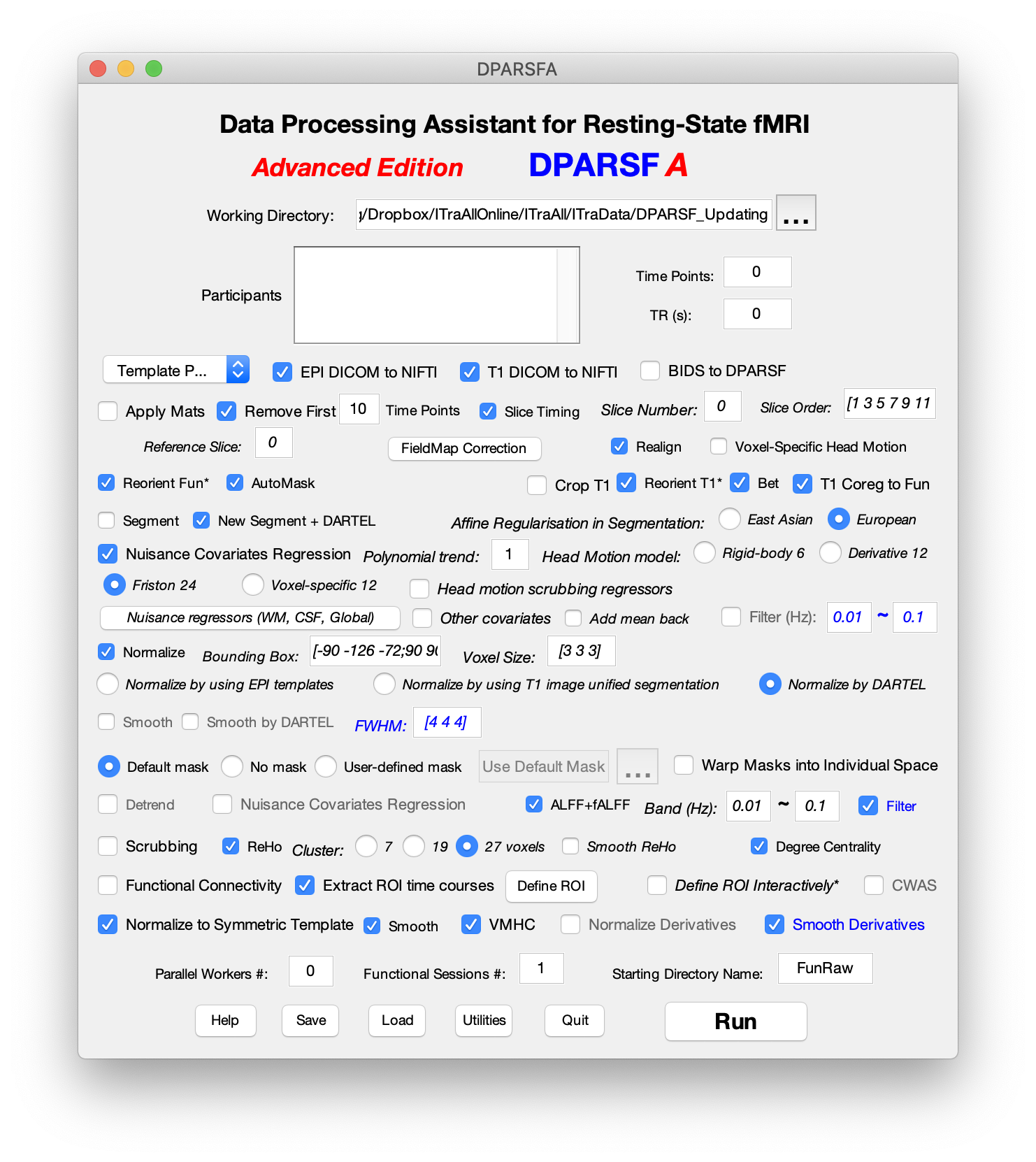
Different Brodmann Atlas in DPABI and REST
- Read more about Different Brodmann Atlas in DPABI and REST
- 8 comments
- Log in or register to post comments
Dear experts,
I found that Brodmann Atlas was used in DPABI Viewer and REST Slice Viewer. In REST, there are 48 areas in the Brodmann Atlas, whereas there are only 41 areas in DPABI. I wonder if there are any other differences between these two atlas? And which one is recommanded?
Thanks in advance!
Vincent

.png)
.png)