Alphasim and GRF correction
- Read more about Alphasim and GRF correction
- 3 comments
- Log in or register to post comments
Dear experts,
Dear experts,
Dear experts,
Hi!
It could be a silly question, sorry!!! but I'm a rookie in this field!
I was doing a DPARSF with hdr img files functional and structural. with my data subjects and with the data from one of my lab colleagues.
When I was at the step of corregister & reslice I realized that his data had a different matrix dimensions (only for the structural one).
I have 256 x 256 and he has a lower one 182 x 246.
So, here is my doubt, I can resample his structural data to my matrix dimensions? I don't have his raw data.
Dear Dr. Yan,
I am running DPARSF v2.3 on fcon_1000/Beijing_zang data. As suggested by the link http://www.rfmri.org/DownloadedReorientMats, I put the folder “DownloadedReorientMats” (DownloadedReorientMats_FCP/Beijing/) under my workding directory and set up DPARSF as follows.
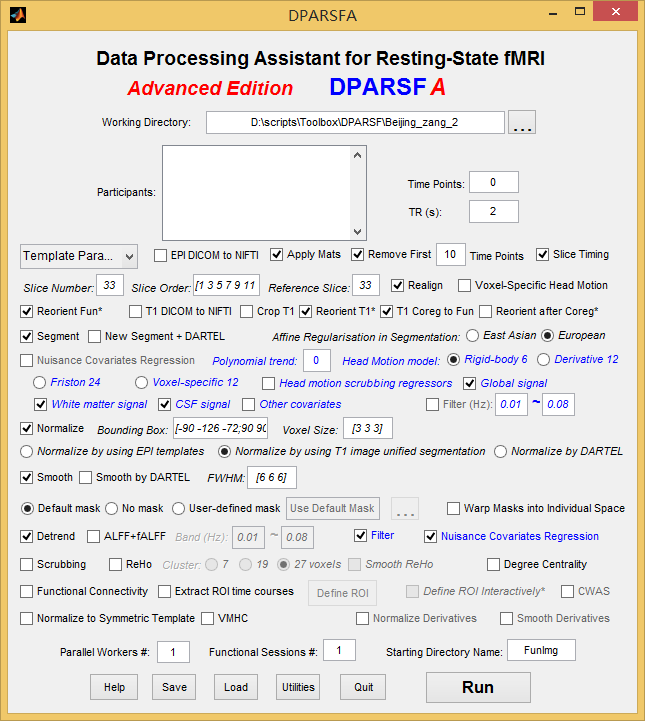
Hi again,
I was trying with DPARSFA running just the FC adding masks rois but it didn't work properly.
When I performed the whole analyisis it worked perfectly, now running just FC in DPARSFA when I perform two test analysis I can see voxels outside the brain.
Where is the mistake?
Thanks in advance!!
Dear experts,
I want to know, if I'm on the right way performing this two test.
I've 2 resting groups and I pretend to compare a specific Zroi files from a previous DPARSF analysis without any kind of mask.
Just one group against the other.
What do you think? It will be ok? or it will be better to perform it with a created or a template mask for each ROI.
Basically what is happening is that immediately after the detrend step and prior to the filter step, the script crashes giving this error
Error using rmdir
No directories were removed.
Error in DPARSF_run (line 998)
rmdir([AutoDataProcessParameter.SubjectID(i), '_detrend']) ;
I have tried multiple things such as restarting from the beginning and ticking and unticking the delete detrend option. Any suggestions would be greatly appreciated.
Dear DPARSFers,
Could you please confirm if the values that I am enering are correct?
Dear experts,
I am getting an error in the detrend stage of DPARSF. I could reproduce this error many times (I didn't click any button nor interacted in any way with the software). Note that I also get some warnings that I don't know if I should be worried about.
I am running this on Matlab R2007b on a MacOSX 10.6.
Thanks in advance for your help!
Best,
Miguel
Hi Chao-Gan,
I am reading your standardization paper and would like to implement it going forward.
However, as far as I can see, the processing stream calls for slice timing correction, motion correction, and nuisance covariate analyses (using Friston24, CSF, and WM, but without GSR). Wouldn't this give a mean of zero for the FunImgRC files? If so, where does the mean come from in the group level analysis? Do I just take the mean and SD of the FunImgR images and use those?
Best,
Matt